# Read subsetted data from online file - make sure there are no spaces
gtex = read_tsv('https://tinyurl.com/342rhdc2')
# Check number of rows
nrow(gtex)[1] 389922This is a subset of the Genotype Tissue Expression (GTEx) dataset
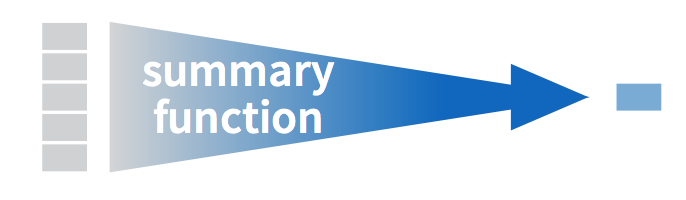
summarize() boils down the data frame according to the conditions it gets. In this case, it creates a data frame with a single column called blood_avg that contains the mean of the Blood columnmutate(), the name on the left of the = is something you make up that you would like the new column to be named.mutate() transforms columns into new columns of the same length, but summarize() collapses down the data frame into a single row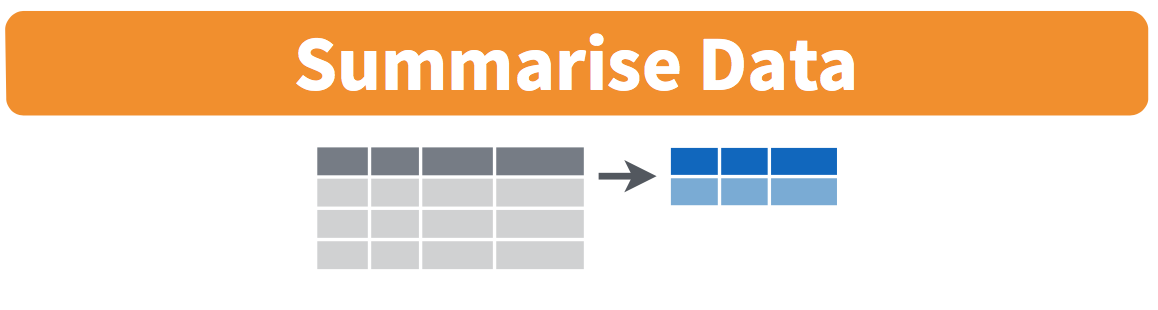
Find the average, maximum, and minimum expressions of the REN gene in blood. Hint: filter and summarize.
# A tibble: 4,999 × 2
Gene max_blood
<chr> <dbl>
1 A2ML1 2.08
2 A3GALT2 2.77
3 A4GALT 2.78
4 AAMDC NA
5 AANAT 1.71
6 AAR2 2.52
7 AARSD1 1.89
8 AB019441.29 2.31
9 ABC7-42389800N19.1 1.98
10 ABCA5 2.3
# ℹ 4,989 more rowsgroup_by() is a helper function that “groups” the data according to the unique values in the column(s) it gets passed.summarize() works the same as before, except now it returns as many rows as there are groups in the data
gtex |>
filter(!is.na(Blood) & !is.na(Lung)) |>
group_by(
pos_blood = Blood>0,
pos_lung = Lung>0
) |>
summarize(mean_liver = mean(Liver, na.rm=T))# A tibble: 4 × 3
# Groups: pos_blood [2]
pos_blood pos_lung mean_liver
<lgl> <lgl> <dbl>
1 FALSE FALSE -0.106
2 FALSE TRUE 0.0145
3 TRUE FALSE -0.0141
4 TRUE TRUE 0.109 n() function counts the number of rows in each group:# A tibble: 4,999 × 2
Gene how_many
<chr> <int>
1 A2ML1 78
2 A3GALT2 78
3 A4GALT 78
4 AAMDC 77
5 AANAT 78
6 AAR2 78
7 AARSD1 78
8 AB019441.29 78
9 ABC7-42389800N19.1 78
10 ABCA5 78
# ℹ 4,989 more rowscount(), which is just a shorthand for the same thingIgnoring NAs, what are the highest and lowest liver expression values seen for each gene in the gtex dataset?
What steps should you take to solve this problem? When you have a question that asks something about “for each …” that usually indicates that you need to group_by() the data by whatever thing that is. When you are asking about a summary measure (like a mean, max etc.), that usually indicates the use of summarize(). In this problem, what column(s) are you grouping by? What summaries of what columns are you computing?
Now that you have a structure, write the code to implement it and solve the problem.
Before continuing, run this code to reformat your data and store it as a new data frame gtex_tidy (we’ll see how to do this later today):
Have a look at the dataframe you created. Use it to recreate this plot:

The max_expression variable in the x-axis of the plot indicates the maximum expression across all samples (in whatever grouping we are looking at).
It’s helpful to think backwards from the output you want. First outline the ggplot code that would generate the given plot. What does the dataset need to look like that is going into ggplot in order to get the plot shown here? How can we get from gtex_tidy to that data?
filter() is aware of grouping. When used on a grouped dataset, it applies the filtering condition separately in each group# A tibble: 79 × 3
# Groups: Ind [78]
Ind Gene Lung
<chr> <chr> <dbl>
1 GTEX-11TUW AC007743.1 4.32
2 GTEX-147F4 ACRV1 5.4
3 GTEX-YFC4 ALOXE3 7.5
4 GTEX-ZPU1 ANKDD1B 4.12
5 GTEX-X4EP AP001610.5 6.59
6 GTEX-1GN2E ATF4P3 6.95
7 GTEX-1LGRB CASP12 3.66
8 GTEX-1E2YA COLGALT1 6.96
9 GTEX-17HGU CTAG2 7.4
10 GTEX-14E1K CTD-2525I3.5 5.9
# ℹ 69 more rowsgtex |>
group_by(Gene) |>
mutate(rank = rank(-Blood)) |>
select(Gene, Ind, rank, Blood) |>
filter(rank <= 3) |>
arrange(Gene, rank)# A tibble: 14,867 × 4
# Groups: Gene [4,999]
Gene Ind rank Blood
<chr> <chr> <dbl> <dbl>
1 A2ML1 GTEX-1A8FM 1 2.08
2 A2ML1 GTEX-WY7C 2 1.73
3 A2ML1 GTEX-ZTPG 3 1.11
4 A3GALT2 GTEX-1AX9I 1 2.77
5 A3GALT2 GTEX-14XAO 2 1.54
6 A3GALT2 GTEX-1B933 3 1.41
7 A4GALT GTEX-12696 1 2.78
8 A4GALT GTEX-18A6Q 2 2.66
9 A4GALT GTEX-11DXZ 3 2.02
10 AAMDC GTEX-14XAO 1 2.32
# ℹ 14,857 more rowsmutate is used on a grouped dataset, it applies the mutation separately in each groupgtex |>
mutate(rank = rank(-Blood)) |>
select(Gene, Ind, rank, Blood) |>
filter(rank <= 3) |>
arrange(Gene, rank)# A tibble: 3 × 4
Gene Ind rank Blood
<chr> <chr> <dbl> <dbl>
1 DNASE2B GTEX-12696 3 14.4
2 KLK3 GTEX-147F4 2 15.7
3 REN GTEX-U8XE 1 18.9group_by, the ranking is done overall across all genes.Create a dataset that shows which gene has the lowest expression in each person’s heart tissue
# A tibble: 3 × 8
tissue `2011` `2012` `2013` `2014` `2015` `2016` `2017`
<chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 Adipose Tissue 56 107 243 206 84 134 2
2 Adrenal Gland 28 41 84 65 20 31 0
3 Bladder 2 18 0 1 0 0 0# A tibble: 3 × 8
tissue `2011` `2012` `2013` `2014` `2015` `2016` `2017`
<chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 Adipose Tissue 56 107 243 206 84 134 2
2 Adrenal Gland 28 41 84 65 20 31 0
3 Bladder 2 18 0 1 0 0 0# A tibble: 6 × 3
tissue year count
<chr> <chr> <dbl>
1 Adipose Tissue 2011 56
2 Adipose Tissue 2012 107
3 Adipose Tissue 2013 243
4 Adipose Tissue 2014 206
5 Adipose Tissue 2015 84
6 Adipose Tissue 2016 134This data is tidy. Tidy data follows three precepts:
In our example, each of the observations are different groups of samples, each of which has an associated tissue, year, and count. These are the variables that are associated with the groups of samples.
Tidy data is easy to work with.
tidyr::pivot_longer() is the function you will most often want to use to tidy your datatidyr::pivot_longer() is the function you will most often want to use to tidy your data# A tibble: 2 × 3
tissue year count
<chr> <chr> <dbl>
1 Adipose Tissue 2011 56
2 Adipose Tissue 2012 107# A tibble: 4 × 3
mouse weight_before weight_after
<dbl> <dbl> <dbl>
1 1 10.5 9.92
2 2 5.92 7.28
3 3 8.03 14.1
4 4 10.5 9.99# A tibble: 8 × 3
mouse time weight
<dbl> <chr> <dbl>
1 1 before 10.5
2 1 after 9.92
3 2 before 5.92
4 2 after 7.28
5 3 before 8.03
6 3 after 14.1
7 4 before 10.5
8 4 after 9.99pivot_wider():I have a dataset that records the pollution levels (in ppm) in three cities across five months:
# A tibble: 15 × 3
city month smoke_ppm
<chr> <chr> <dbl>
1 SF Jan 14.4
2 SF Feb 39.4
3 SF Mar 20.4
4 SF Apr 44.2
5 SF May 47.0
6 LA Jan 2.28
7 LA Feb 26.4
8 LA Mar 44.6
9 LA Apr 27.6
10 LA May 22.8
11 NY Jan 47.8
12 NY Feb 22.7
13 NY Mar 33.9
14 NY Apr 28.6
15 NY May 5.15Use a pivot and mutate to compute the difference in pollution levels between SF and LA across all 5 months. The output should look like this:
# A tibble: 5 × 4
month SF LA SF_LA_diff
<chr> <dbl> <dbl> <dbl>
1 Jan 14.4 2.28 12.1
2 Feb 39.4 26.4 13.0
3 Mar 20.4 44.6 -24.2
4 Apr 44.2 27.6 16.6
5 May 47.0 22.8 24.2Use the GTEX data to reproduce the following plot:
The individuals and genes of interest are c('GTEX-11GSP', 'GTEX-11DXZ') and c('A2ML1', 'A3GALT2', 'A4GALT'), respectively.
Think backwards: what do the data need to look like to make this plot? How do we pare down and reformat gtex so that it looks like what we need?
Use the GTEX data to make the following table:
[1] "Number of missing tissues:"# A tibble: 2 × 4
# Groups: Ind [2]
Ind A2ML1 A3GALT2 A4GALT
<chr> <int> <int> <int>
1 GTEX-11DXZ 1 0 0
2 GTEX-11GSP 0 0 0The numbers in the table are the number of tissues in each individual for which the gene in question was missing.
# A tibble: 1 × 6
subject_id sample_id batch_id center_id tissue rin_score
<chr> <chr> <chr> <chr> <chr> <dbl>
1 GTEX-11DXZ 0003-SM-58Q7X BP-39216 B1 Blood NA# A tibble: 1 × 4
subject_id sex age death
<chr> <chr> <chr> <chr>
1 GTEX-11DXZ male 50-59 ventilatorsubject data are referenced in the sample data, and the batches referenced in the sample data are in the batch data. The sample ids from the sample data are used for accessing expression data.subject connects to sample via a single variable, subject_id.sample connects to batch through the batch_id variable.# A tibble: 312 × 9
subject_id sample_id batch_id center_id tissue rin_score sex age death
<chr> <chr> <chr> <chr> <chr> <dbl> <chr> <chr> <chr>
1 GTEX-11DXZ 0003-SM-58Q… BP-39216 B1 Blood NA male 50-59 vent…
2 GTEX-11DXZ 0126-SM-5EG… BP-44460 B1 Liver 7.9 male 50-59 vent…
3 GTEX-11DXZ 0326-SM-5EG… BP-44460 B1 Heart 8.3 male 50-59 vent…
4 GTEX-11DXZ 0726-SM-5N9… BP-43956 B1 Lung 7.8 male 50-59 vent…
5 GTEX-11GSP 0004-SM-58Q… BP-39412 B1 Blood NA fema… 60-69 sudd…
6 GTEX-11GSP 0626-SM-598… BP-44902 B1 Liver 6.2 fema… 60-69 sudd…
7 GTEX-11GSP 0726-SM-598… BP-44902 B1 Lung 6.9 fema… 60-69 sudd…
8 GTEX-11GSP 1226-SM-598… BP-44902 B1 Heart 7.9 fema… 60-69 sudd…
9 GTEX-11NUK 0004-SM-58Q… BP-39723 B1 Blood NA male 50-59 sudd…
10 GTEX-11NUK 0826-SM-5HL… BP-43730 B1 Lung 7.4 male 50-59 sudd…
# ℹ 302 more rows
# A tibble: 2 × 3
name plays band
<chr> <chr> <chr>
1 John guitar Beatles
2 Paul bass Beatles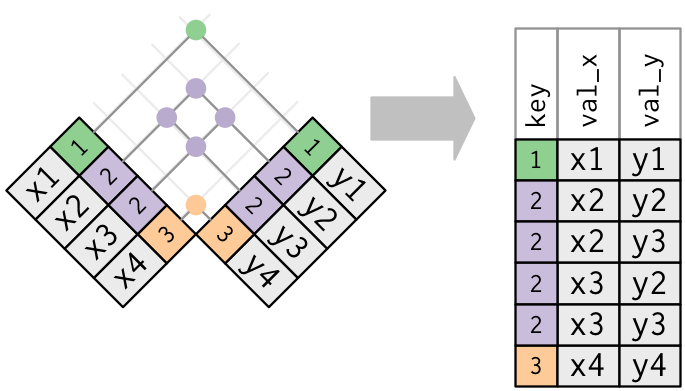
Error in `inner_join()`:
! Join columns in `y` must be present in the data.
✖ Problem with `center_id`.# A tibble: 2 × 6
Gene Ind Blood Heart Lung Liver
<chr> <chr> <dbl> <dbl> <dbl> <dbl>
1 A2ML1 GTEX-11DXZ -0.14 -1.08 NA -0.66
2 A2ML1 GTEX-11GSP -0.5 0.53 0.76 -0.1 # A tibble: 2 × 4
subject_id sex age death
<chr> <chr> <chr> <chr>
1 GTEX-11DXZ male 50-59 ventilator
2 GTEX-11GSP female 60-69 sudden but natural causes# A tibble: 5 × 9
Gene Ind Blood Heart Lung Liver sex age death
<chr> <chr> <dbl> <dbl> <dbl> <dbl> <chr> <chr> <chr>
1 A2ML1 GTEX-11DXZ -0.14 -1.08 NA -0.66 male 50-59 ventilator
2 A2ML1 GTEX-11GSP -0.5 0.53 0.76 -0.1 female 60-69 sudden but natural caus…
3 A2ML1 GTEX-11NUK -0.08 -0.4 -0.26 -0.13 male 50-59 sudden but natural caus…
4 A2ML1 GTEX-11NV4 -0.37 0.11 -0.42 -0.61 male 60-69 sudden but natural caus…
5 A2ML1 GTEX-11TT1 0.3 -1.11 0.59 -0.12 male 20-29 ventilator Note that the first key (Ind) corresponds to the first data frame (gtex) and the second key (subject_id) corresponds to the second data frame (gtex_subjects).
How does the average A2ML1 expression in lung tissue compare between females vs. males? To answer this question let’s break it down:
gtex to only include those rows.gtex_subjects data frame.Write the code to execute these steps.
Use joins to find the samples collected in 2015 with blood expression greater than 3 of “KRT19” in males. Start with the batch_data_year; this data has an extra extracted column with the year.
batch_data_year =
gtex_batches |>
mutate(
batch_date = lubridate::mdy(batch_date),
year = lubridate::year(batch_date)
)
head(batch_data_year, 2)# A tibble: 2 × 4
batch_id batch_type batch_date year
<chr> <chr> <date> <dbl>
1 BP-38516 DNA isolation_Whole Blood_QIAGEN Puregene (Manual) 2013-05-02 2013
2 BP-42319 RNA isolation_PAXgene Tissue miRNA 2013-08-14 2013Start by figuring out what other data frame(s) you have to join to. Consider also what other operations you must do to pick out the data of interest and in what order (if it matters).
Another common way to combine two data frames is bind_rows (or bind_cols). Read the documentation for those functions and compare to what you know about joins. What is fundamentally different about binding (concatenating) vs. joining?
When would you do one vs. the other?
Let’s read in some more data
# A tibble: 2 × 4
tissue month year tiss_samples
<chr> <dbl> <dbl> <dbl>
1 Adipose Tissue 1 2012 13
2 Adipose Tissue 1 2013 4# A tibble: 2 × 3
month year num_samples
<dbl> <dbl> <dbl>
1 5 2011 20
2 6 2011 44# A tibble: 5 × 5
tissue month year tiss_samples num_samples
<chr> <dbl> <dbl> <dbl> <dbl>
1 Adipose Tissue 1 2012 13 208
2 Adipose Tissue 1 2013 4 64
3 Adipose Tissue 1 2014 52 662
4 Adipose Tissue 1 2015 20 263
5 Adipose Tissue 1 2016 7 107