R Programming
Learning Goals:
- save values to variables
- find and call R functions with multiple arguments by position and name
- recognize and index vectors and lists
- recognize, import, and inspect data frames
- issue commands to R using the Rstudio script pane
Programming Basics
- We’ve seen code like
- We know this reads a .csv from a file and creates something called a “data frame”
- We’ve been using this data frame in code like
- But what does this syntax really mean? Is it useful outside of making plots?
Assignment
Assignment
- To do complex computations, we need to be able to give names to things.
- This code assigns the result of running
read_csv("https://tinyurl.com/cjkuecnc")to the namegene - You can do this with any values and/or functions
Variables
- When R sees the name of a variable, it uses the stored value of that variable in the calculation.
- We can break complex calculations into named parts. This is a simple, but very useful kind of abstraction.

Two ways to assign
In R, there are (unfortunately) two assignment operators. They have subtly different meanings (more details later).
<-requires that you type two characters but better captures spirit of assignmnet=is easier to type but incorrectly suggests mathematical equality- You will see both used throughout R and user code.
Assignment has no undo
- If you assign to a name with an existing value, that value is overwritten.
- There is no way to undo an assignment, so be careful in reusing variable names.
Naming variables
- It is important to pick meaningful variable names.
- Names can be too short, so don’t use
xandyeverywhere. - Names can be too long (
Main.database.first.object.header.length). - Avoid silly names.
- Pick names that will make sense to someone else (including the person you will be in six months).
- ADVANCED: See
?make.namesfor the complete rules on what can be a name.
There are different conventions for constructing compound names. Warning: disputes over the right way to do this can get heated.
- To be consistent with the packages we will use, I recommend snake_case where you separate lowercase words with _
- Note that R itself uses several of these conventions.
- One of these won’t work. Which one and why?
Error: object 'A' not found- R cares about upper and lower case in names.
- names can’t start with numbers
foris a reserved word in R. (It is used in loop control.)- ADVANCED: see
?Reservedfor the complete rules.
Exercise: birth year
- Make a variable that represents the age you will be at the end of this year
- Make a variable that represents the current year
- Use them to compute the year of your birth and save that as a variable
- Print the value of that variable
Assignment and Reference
- What do you observe?


Functions
Calling functions
- To call a function, type the function name, then the argument or arguments in parentheses. (Use a comma to separate the arguments, if more than one.)
Arguments
- Functions transform inputs to outputs
- internally, however, they have an environment just like the one you see in your workspace
- when you call a function, you tell it how to connect the variables in your environment to the ones it expects to have so that it can do its job
- the names the function calls these inputs inside itself will be different than what you call them on the outside
aes(x=EIF3L, y=VAPA)
Named and positional arguments
- Arguments can be supplied by name using the syntax variable
=value. - you can see the names of the arguments in the help page for each function
- When using names, the order of the named arguments does not matter.
Optional arguments
- Many R functions have arguments that you don’t always have to specify. For example:
n_maxtellsread_csv()to only read the first 10 rows of the dataset.- If you don’t specify it, it defaults to infinity (i.e. R reads until there are no more lines in the file).
Exercise [together]
Why does this code generate errors?
Exercise [together]
Functions, assignment, and reference
- What do you observe?

- functions generally do not affect the variables you pass to them (
xremains the same aftersqrt(x))
Vectors
Repetitive calculations
Let’s say I have these variables and I want to add 1 to all of them and save the result.
This does the trick but it’s a lot of copy-paste
Vectors
- Vectors solve the problem
- A vector is a one-dimensional sequence of zero or more values
- Vectors are created by wrapping the values separated by commas with the
c()function, which is short for “combine” - Many R functions and operators (like
+) automatically work with multi-element vector arguments.
Elementwise operations
- This multiplies each element of
c(1,2,3)by the corresponding element ofc(4,5,6)
- Many basic R functions operate on multi-element vectors as easily as on vectors containing a single number.
Summaries
- some R functions take vectors as arguments and summarize them instead of applying elementwise
Exercise: subtract the mean
A particular class has two quizzes which are taken by the same three students. Thier scores are below:
- Write code to see if the quiz 1 average is higher than or lower than the quiz 2 average
- Are students improving? Subtract the quiz 1 scores from the quiz 2 scores and take the average of the resulting differences to find out.
Exercise: a vector of variables [together]
- Predict the output of the following code:
Ranges
[1] 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25
[26] 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50- The colon
:is a handy shortcut to create a vector that is a sequence of integers from the first number to the second number (inclusive). - Ranges can go the other way too and include negative numbers, e.g.
5:-5 - Long vectors wrap around. (Your screen may have a different width than what is shown here.)
Indexing
[1] "a"[1] "b" "c" "d"[1] "c" "a"[1] "a" "a" "a" "a" "a" "a" "d"- Indexing returns a subsequence of the vector. It does not change the original vector. Assign the result to a new variable to save it if you neeed it later.
- R starts counting vector indices from 1.
- You can index using a multi-element index vector.
- You can repeat index positions
Exercise: reading vector code
What does this code do?
- read inside out: first figure out what
length(x)does, then think about what the output oflength(x):1should do, and then finallyx[length(x):1]
Indexed Assignment
- you can assign into an indexed position
- or multiple
Data Types
Strings
- text data in R is called a “string”
- when using data that is text in R, you have to refer to it using quotation marks (why?)
- you can have a vector of strings, and functions can operate on these too:
Factors
- factors represent categorical data
[1] "spring" "summer" "fall" "winter"[1] spring summer fall winter
Levels: spring summer fall winter- this is useful to tightly control data and prevent accidents
Logicals
- logical vectors can only be
TRUEorFALSE - we’ll see more about this later
Coercion
- If you try to do something to a vector of the wrong data type, R will often do its best to “make it work” by converting to another type
Error in numbers + 2: non-numeric argument to binary operator- this is a frequent source of unexpected errors!
Exercise: data types [together]
What types are each of the following vectors? Are they all fundamentally the same, or are they different?
Which of these lines of code will run and which will produce an error?
NA
- R has a special value that represents missing data- it’s called
NA
- NA can appear anywhere that R would expect some other kind of data
- NA usually ruins computations:
- The result makes sense because if I don’t know what I’m adding together, I don’t know the result either
- some functions have options to ignore the missing values in vectors:
Lists
Lists
- A
listis like an atomic vector, except the elements don’t have to be the same type of thing
a_vector = c(1,2,4,5)
maybe_a_vector = c(1,2,"hello",5,TRUE)
maybe_a_vector # R converted all of these things to strings![1] "1" "2" "hello" "5" "TRUE" - You make them with list() and you can index them like vectors
- Anything can go in lists, including vectors, other lists, data frames, etc.
- In fact, a data frame (or tibble) is actually just a list of named column vectors with an enforced constraint that all of the vectors have to be of the same length. That’s why the
df$colsyntax works for data frames.
Getting elements from a list
- You can also name the elements in a list
- and then retrieve elements by name or position
Examining lists
- Use
str()to dig into nested lists and other complicated objects
List of 12
$ coefficients : Named num [1:11] 79.048 -2.063 8.204 0.439 -4.619 ...
..- attr(*, "names")= chr [1:11] "(Intercept)" "mpg" "cyl" "disp" ...
$ residuals : Named num [1:32] -38.68 -30.63 13.01 -15.75 -8.22 ...
..- attr(*, "names")= chr [1:32] "Mazda RX4" "Mazda RX4 Wag" "Datsun 710" "Hornet 4 Drive" ...
$ effects : Named num [1:32] -829.8 296.3 124.8 -19.6 90.3 ...
..- attr(*, "names")= chr [1:32] "(Intercept)" "mpg" "cyl" "disp" ...
$ rank : int 11
$ fitted.values: Named num [1:32] 149 141 80 126 183 ...
..- attr(*, "names")= chr [1:32] "Mazda RX4" "Mazda RX4 Wag" "Datsun 710" "Hornet 4 Drive" ...
$ assign : int [1:11] 0 1 2 3 4 5 6 7 8 9 ...
$ qr :List of 5
..$ qr : num [1:32, 1:11] -5.657 0.177 0.177 0.177 0.177 ...
.. ..- attr(*, "dimnames")=List of 2
.. .. ..$ : chr [1:32] "Mazda RX4" "Mazda RX4 Wag" "Datsun 710" "Hornet 4 Drive" ...
.. .. ..$ : chr [1:11] "(Intercept)" "mpg" "cyl" "disp" ...
.. ..- attr(*, "assign")= int [1:11] 0 1 2 3 4 5 6 7 8 9 ...
..$ qraux: num [1:11] 1.18 1.02 1.29 1.19 1.05 ...
..$ pivot: int [1:11] 1 2 3 4 5 6 7 8 9 10 ...
..$ tol : num 1e-07
..$ rank : int 11
..- attr(*, "class")= chr "qr"
$ df.residual : int 21
$ xlevels : Named list()
$ call : language lm(formula = hp ~ ., data = mtcars)
$ terms :Classes 'terms', 'formula' language hp ~ mpg + cyl + disp + drat + wt + qsec + vs + am + gear + carb
.. ..- attr(*, "variables")= language list(hp, mpg, cyl, disp, drat, wt, qsec, vs, am, gear, carb)
.. ..- attr(*, "factors")= int [1:11, 1:10] 0 1 0 0 0 0 0 0 0 0 ...
.. .. ..- attr(*, "dimnames")=List of 2
.. .. .. ..$ : chr [1:11] "hp" "mpg" "cyl" "disp" ...
.. .. .. ..$ : chr [1:10] "mpg" "cyl" "disp" "drat" ...
.. ..- attr(*, "term.labels")= chr [1:10] "mpg" "cyl" "disp" "drat" ...
.. ..- attr(*, "order")= int [1:10] 1 1 1 1 1 1 1 1 1 1
.. ..- attr(*, "intercept")= int 1
.. ..- attr(*, "response")= int 1
.. ..- attr(*, ".Environment")=<environment: R_GlobalEnv>
.. ..- attr(*, "predvars")= language list(hp, mpg, cyl, disp, drat, wt, qsec, vs, am, gear, carb)
.. ..- attr(*, "dataClasses")= Named chr [1:11] "numeric" "numeric" "numeric" "numeric" ...
.. .. ..- attr(*, "names")= chr [1:11] "hp" "mpg" "cyl" "disp" ...
$ model :'data.frame': 32 obs. of 11 variables:
..$ hp : num [1:32] 110 110 93 110 175 105 245 62 95 123 ...
..$ mpg : num [1:32] 21 21 22.8 21.4 18.7 18.1 14.3 24.4 22.8 19.2 ...
..$ cyl : num [1:32] 6 6 4 6 8 6 8 4 4 6 ...
..$ disp: num [1:32] 160 160 108 258 360 ...
..$ drat: num [1:32] 3.9 3.9 3.85 3.08 3.15 2.76 3.21 3.69 3.92 3.92 ...
..$ wt : num [1:32] 2.62 2.88 2.32 3.21 3.44 ...
..$ qsec: num [1:32] 16.5 17 18.6 19.4 17 ...
..$ vs : num [1:32] 0 0 1 1 0 1 0 1 1 1 ...
..$ am : num [1:32] 1 1 1 0 0 0 0 0 0 0 ...
..$ gear: num [1:32] 4 4 4 3 3 3 3 4 4 4 ...
..$ carb: num [1:32] 4 4 1 1 2 1 4 2 2 4 ...
..- attr(*, "terms")=Classes 'terms', 'formula' language hp ~ mpg + cyl + disp + drat + wt + qsec + vs + am + gear + carb
.. .. ..- attr(*, "variables")= language list(hp, mpg, cyl, disp, drat, wt, qsec, vs, am, gear, carb)
.. .. ..- attr(*, "factors")= int [1:11, 1:10] 0 1 0 0 0 0 0 0 0 0 ...
.. .. .. ..- attr(*, "dimnames")=List of 2
.. .. .. .. ..$ : chr [1:11] "hp" "mpg" "cyl" "disp" ...
.. .. .. .. ..$ : chr [1:10] "mpg" "cyl" "disp" "drat" ...
.. .. ..- attr(*, "term.labels")= chr [1:10] "mpg" "cyl" "disp" "drat" ...
.. .. ..- attr(*, "order")= int [1:10] 1 1 1 1 1 1 1 1 1 1
.. .. ..- attr(*, "intercept")= int 1
.. .. ..- attr(*, "response")= int 1
.. .. ..- attr(*, ".Environment")=<environment: R_GlobalEnv>
.. .. ..- attr(*, "predvars")= language list(hp, mpg, cyl, disp, drat, wt, qsec, vs, am, gear, carb)
.. .. ..- attr(*, "dataClasses")= Named chr [1:11] "numeric" "numeric" "numeric" "numeric" ...
.. .. .. ..- attr(*, "names")= chr [1:11] "hp" "mpg" "cyl" "disp" ...
- attr(*, "class")= chr "lm"Data Frames
Making data frames
- use
tibble()to make your own data frames from scratch in R
Data frame properties
dim()gives the dimensions of the data frame.ncol()andnrow()give you the number of columns and the number of rows, respectively.
names()gives you the names of the columns (a vector)
glimpse()shows you a lot of information,head()returns the firstnrows
Writing data frames
- after running this, you’ll see a new file called
my_data.csv(or whatever you chose to name it) appear in the specified location on your computer (e.g.Desktop) - you can read and write
.csvfiles in lots of programs (e.g. google sheets) - to read and write other formats look at documentation and use google + chatGPT!
Scripts
Using the script pane
- Writing a series of expressions in the console rapidly gets messy and confusing.
- The console window gets reset when you restart RStudio.
- It is better (and easier) to write expressions and functions in the script pane (upper left), building up your analysis.
- There, you can enter expressions, evaluate them, and save the contents to a .R file for later use.
- Look at the RStudio ``Code’’ menu for some useful keyboard commands.
- Create a script pane: File > New File > R Script
- Put your cursor in the script pane.
- Type:
1:10^2 - Then hit
Command-RETURN(Mac), orCtrl-ENTER(Windows). - That line is copied to the console pane and evaluated.
- You can save the script to a file.
- Explore the RStudio Code menu for other commands.
RStudio Pro-tip: scrolling and multicursors
- You should also be aware of
cmd-<arrow>andalt-<arrow>for moving the cursor (by line and by word) - and
cmd-shift-<arrow>andalt-shift-<arrow>for selecting text (by line and by word) - these also combo with shift (to select) and delete
- RStudio’s script pane supports multi-cursors! Hold
altand drag your mouse up and down - You can also set a keyboard shortcut for
find and add next - These features make it much easier to rename variables, etc.
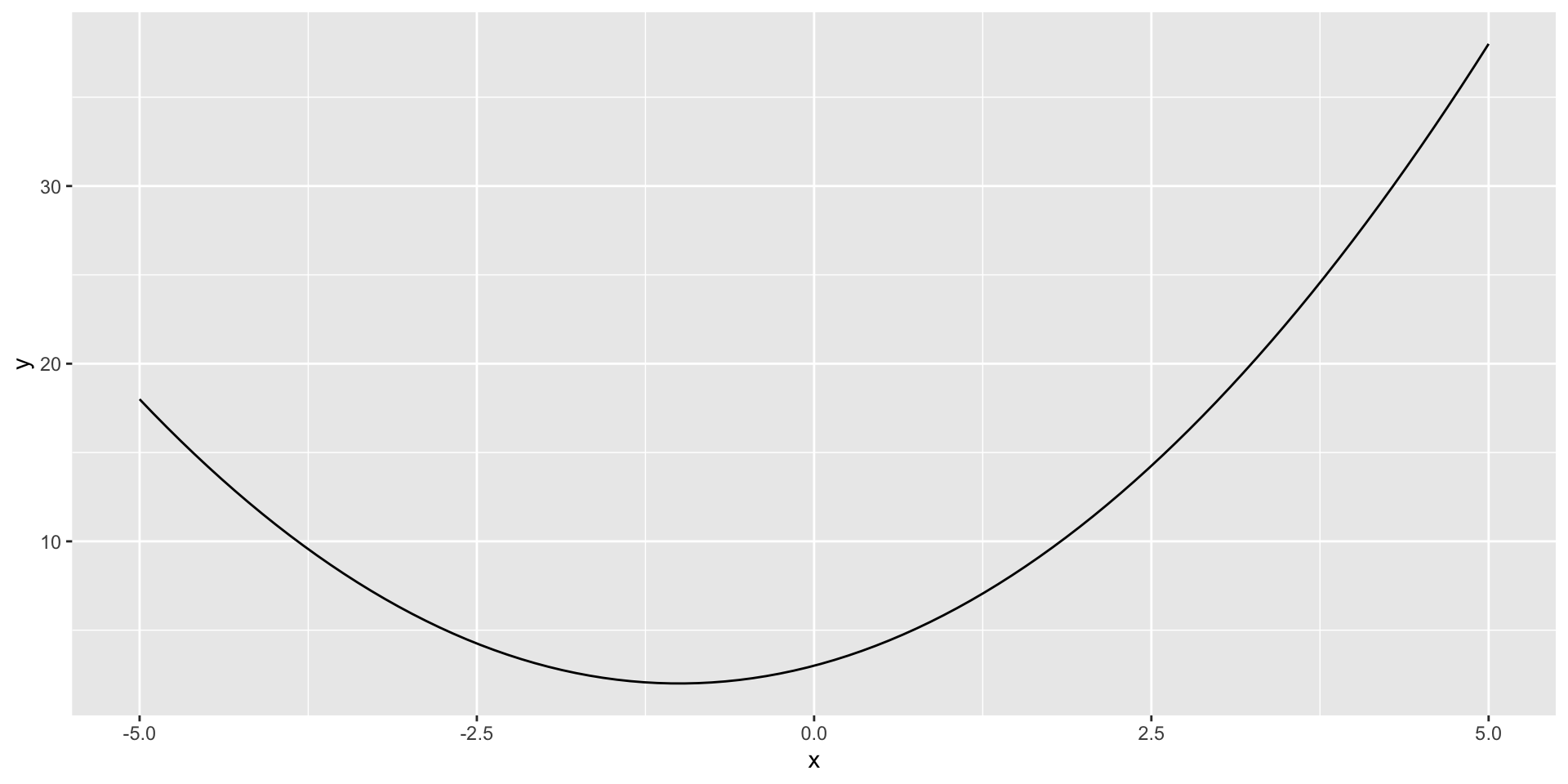
Exercise: Plotting a parabola
Write an R script that starts with:
In the rest of the script, do the following:
- generate an evenly-spaced sequence of 100 values between -5 and 5 (find an R function that does this). Call this
x - generate the corresponding vector of y-values
yby computing the formula \(y = Ax^2 + Bx + C\) elementwise - create a data frame with
xandyas columns - use ggplot to create a line plot of
xvsy
Run your script to see the generated plot. Try changing the values of A, B, and C at the top of the script and re-running to see how the plot changes.
Your result should look like:





Comments
#to start a comment.